Basic cosmology runs
Sampling from a cosmological posterior works the same way as the examples at the beginning of the documentation, except that one usually needs to add a theory code, and possibly some of the cosmological likelihoods presented later.
You can sample or track any parameter that is understood by the theory code in use (or any dynamical redefinition of those). You do not need to modify Cobaya’s source to use new parameters that you have created by modifying CLASS or modifying CAMB, or to create a new cosmological likelihood and track its parameters.
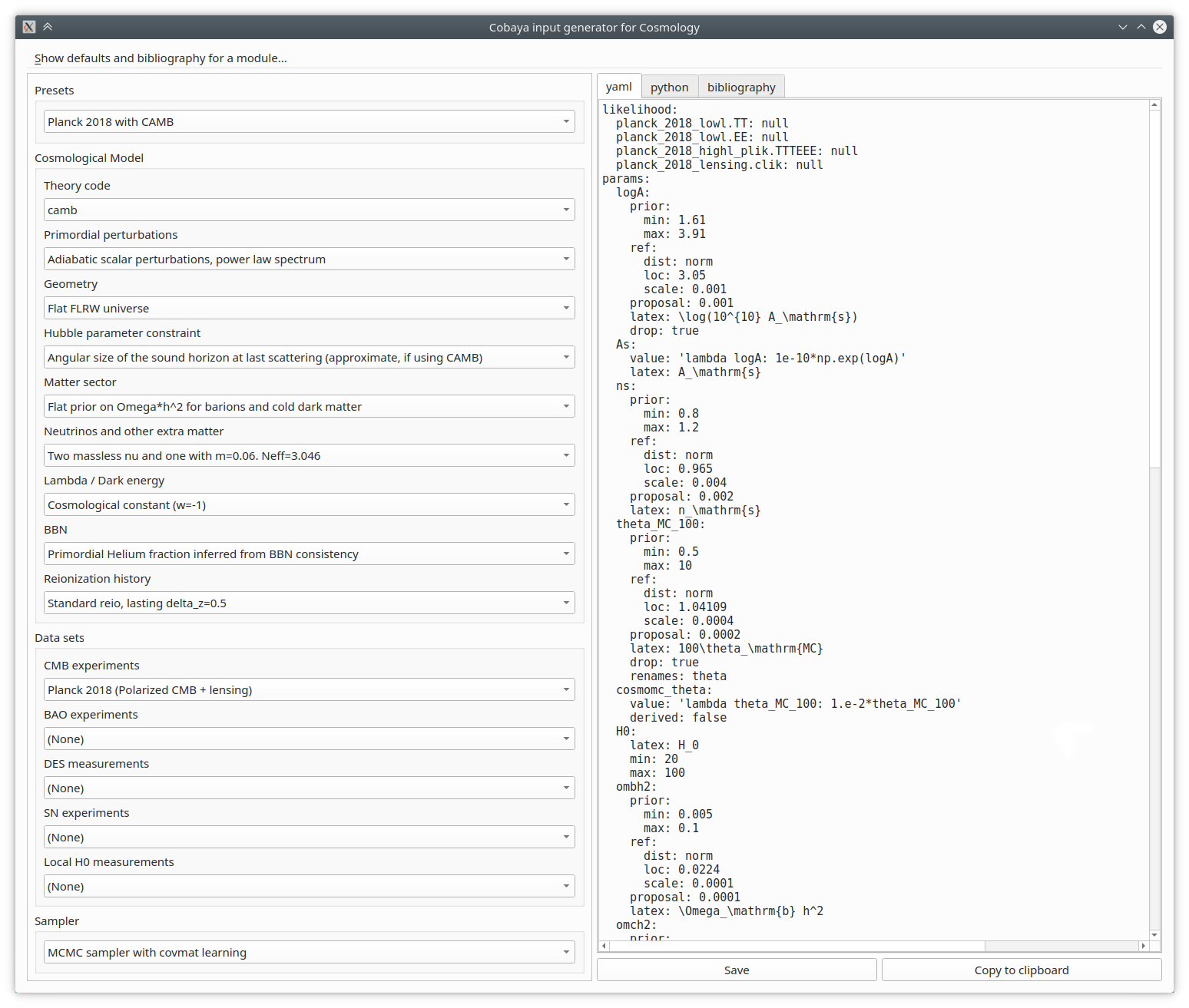
Creating from scratch the input for a realistic cosmological case is quite a bit of work. But to make it simpler, we have created an automatic input generator, that you can run from the shell as:
$ cobaya-cosmo-generator
Note
If
PySideis not installed, this will fail. To fix it:$ python -m pip install PySide6You can also use PySide2. If installing via pip is problematic, the most reliable solution seems to be to make a clean conda-forge environment and use that, e.g. install Anaconda or Miniconda and use the environment created with
$ conda create -n py39forge -c conda-forge python=3.9 scipy pandas matplotlib PyYAML PySide2
Start by choosing a preset, maybe modify some aspects using the options provided, and copy or save the generated input to a file, either in yaml form or as a python dictionary.
The parameter combinations and options included in the input generator are in general well-tested, but they are only suggestions: you can add by hand any parameter that your theory code or likelihood can understand, or modify any setting.
You can add an output prefix if you wish (otherwise, the name of the input file without extension is used). If it contains a folder, e.g. chains/[whatever], that folder will be created if it does not exist.
In general, you do not need to mention the installation path used by cobaya-install (see Installing cosmological codes and data): it will be selected automatically. If that does not work, add packages_path: '/path/to/packages' in the input file, or -p /path/to/packages as a cobaya-run argument.
As an example, here is the input for Planck 2015 base \(\Lambda\mathrm{CDM}\), both for CLASS and CAMB:
Click to toggle CAMB/CLASS
theory:
camb:
extra_args:
halofit_version: mead
bbn_predictor: PArthENoPE_880.2_standard.dat
lens_potential_accuracy: 1
num_massive_neutrinos: 1
nnu: 3.044
theta_H0_range:
- 20
- 100
likelihood:
planck_2018_lowl.TT: null
planck_2018_lowl.EE: null
planck_2018_highl_plik.TTTEEE: null
planck_2018_lensing.clik: null
params:
logA:
prior:
min: 1.61
max: 3.91
ref:
dist: norm
loc: 3.05
scale: 0.001
proposal: 0.001
latex: \log(10^{10} A_\mathrm{s})
drop: true
As:
value: 'lambda logA: 1e-10*np.exp(logA)'
latex: A_\mathrm{s}
ns:
prior:
min: 0.8
max: 1.2
ref:
dist: norm
loc: 0.965
scale: 0.004
proposal: 0.002
latex: n_\mathrm{s}
theta_MC_100:
prior:
min: 0.5
max: 10
ref:
dist: norm
loc: 1.04109
scale: 0.0004
proposal: 0.0002
latex: 100\theta_\mathrm{MC}
drop: true
renames: theta
cosmomc_theta:
value: 'lambda theta_MC_100: 1.e-2*theta_MC_100'
derived: false
H0:
latex: H_0
min: 20
max: 100
ombh2:
prior:
min: 0.005
max: 0.1
ref:
dist: norm
loc: 0.0224
scale: 0.0001
proposal: 0.0001
latex: \Omega_\mathrm{b} h^2
omch2:
prior:
min: 0.001
max: 0.99
ref:
dist: norm
loc: 0.12
scale: 0.001
proposal: 0.0005
latex: \Omega_\mathrm{c} h^2
omegam:
latex: \Omega_\mathrm{m}
omegamh2:
derived: 'lambda omegam, H0: omegam*(H0/100)**2'
latex: \Omega_\mathrm{m} h^2
mnu: 0.06
omega_de:
latex: \Omega_\Lambda
YHe:
latex: Y_\mathrm{P}
Y_p:
latex: Y_P^\mathrm{BBN}
DHBBN:
derived: 'lambda DH: 10**5*DH'
latex: 10^5 \mathrm{D}/\mathrm{H}
tau:
prior:
min: 0.01
max: 0.8
ref:
dist: norm
loc: 0.055
scale: 0.006
proposal: 0.003
latex: \tau_\mathrm{reio}
zrei:
latex: z_\mathrm{re}
sigma8:
latex: \sigma_8
s8h5:
derived: 'lambda sigma8, H0: sigma8*(H0*1e-2)**(-0.5)'
latex: \sigma_8/h^{0.5}
s8omegamp5:
derived: 'lambda sigma8, omegam: sigma8*omegam**0.5'
latex: \sigma_8 \Omega_\mathrm{m}^{0.5}
s8omegamp25:
derived: 'lambda sigma8, omegam: sigma8*omegam**0.25'
latex: \sigma_8 \Omega_\mathrm{m}^{0.25}
A:
derived: 'lambda As: 1e9*As'
latex: 10^9 A_\mathrm{s}
clamp:
derived: 'lambda As, tau: 1e9*As*np.exp(-2*tau)'
latex: 10^9 A_\mathrm{s} e^{-2\tau}
age:
latex: '{\rm{Age}}/\mathrm{Gyr}'
rdrag:
latex: r_\mathrm{drag}
sampler:
mcmc:
drag: true
oversample_power: 0.4
proposal_scale: 1.9
covmat: auto
Rminus1_stop: 0.01
Rminus1_cl_stop: 0.2
theory:
classy:
extra_args:
non linear: hmcode
nonlinear_min_k_max: 20
N_ncdm: 1
N_ur: 2.0328
likelihood:
planck_2018_lowl.TT: null
planck_2018_lowl.EE: null
planck_2018_highl_plik.TTTEEE: null
planck_2018_lensing.clik: null
params:
logA:
prior:
min: 1.61
max: 3.91
ref:
dist: norm
loc: 3.05
scale: 0.001
proposal: 0.001
latex: \log(10^{10} A_\mathrm{s})
drop: true
A_s:
value: 'lambda logA: 1e-10*np.exp(logA)'
latex: A_\mathrm{s}
n_s:
prior:
min: 0.8
max: 1.2
ref:
dist: norm
loc: 0.965
scale: 0.004
proposal: 0.002
latex: n_\mathrm{s}
theta_s_100:
prior:
min: 0.5
max: 10
ref:
dist: norm
loc: 1.0416
scale: 0.0004
proposal: 0.0002
latex: 100\theta_\mathrm{s}
H0:
latex: H_0
omega_b:
prior:
min: 0.005
max: 0.1
ref:
dist: norm
loc: 0.0224
scale: 0.0001
proposal: 0.0001
latex: \Omega_\mathrm{b} h^2
omega_cdm:
prior:
min: 0.001
max: 0.99
ref:
dist: norm
loc: 0.12
scale: 0.001
proposal: 0.0005
latex: \Omega_\mathrm{c} h^2
Omega_m:
latex: \Omega_\mathrm{m}
omegamh2:
derived: 'lambda Omega_m, H0: Omega_m*(H0/100)**2'
latex: \Omega_\mathrm{m} h^2
m_ncdm:
value: 0.06
renames: mnu
Omega_Lambda:
latex: \Omega_\Lambda
YHe:
latex: Y_\mathrm{P}
tau_reio:
prior:
min: 0.01
max: 0.8
ref:
dist: norm
loc: 0.055
scale: 0.006
proposal: 0.003
latex: \tau_\mathrm{reio}
z_reio:
latex: z_\mathrm{re}
sigma8:
latex: \sigma_8
s8h5:
derived: 'lambda sigma8, H0: sigma8*(H0*1e-2)**(-0.5)'
latex: \sigma_8/h^{0.5}
s8omegamp5:
derived: 'lambda sigma8, Omega_m: sigma8*Omega_m**0.5'
latex: \sigma_8 \Omega_\mathrm{m}^{0.5}
s8omegamp25:
derived: 'lambda sigma8, Omega_m: sigma8*Omega_m**0.25'
latex: \sigma_8 \Omega_\mathrm{m}^{0.25}
A:
derived: 'lambda A_s: 1e9*A_s'
latex: 10^9 A_\mathrm{s}
clamp:
derived: 'lambda A_s, tau_reio: 1e9*A_s*np.exp(-2*tau_reio)'
latex: 10^9 A_\mathrm{s} e^{-2\tau}
age:
latex: '{\rm{Age}}/\mathrm{Gyr}'
rs_drag:
latex: r_\mathrm{drag}
sampler:
mcmc:
drag: true
oversample_power: 0.4
proposal_scale: 1.9
covmat: auto
Rminus1_stop: 0.01
Rminus1_cl_stop: 0.2
Note
Note that Planck likelihood parameters (or nuisance parameters) do not appear in the input: they are included automatically at run time. The same goes for all internal likelihoods (i.e. those listed below in the table of contents).
You can still add them to the input, if you want to redefine any of their properties (its prior, label, etc.). See Changing and redefining parameters; inheritance.
Save the input generated to a file and run it with cobaya-run [your_input_file_name.yaml]. This will create output files as explained here, and, after some time, you should be able to run getdist-gui to generate some plots.
Note
You may want to start with a test run, adding --test to cobaya-run (run without MPI). It will initialise all components (cosmological theory code and likelihoods, and the sampler) and exit.
Typical running times for MCMC when using computationally heavy likelihoods (e.g. those involving \(C_\ell\), or non-linear \(P(k,z)\) for several redshifts) are ~10 hours running 4 MPI processes with 4 OpenMP threads per process, provided that the initial covariance matrix is a good approximation to the one of the real posterior (Cobaya tries to select it automatically from a database; check the [mcmc] output towards the top to see if it succeeded), or a few hours on top of that if the initial covariance matrix is not a good approximation.
It is much harder to provide typical PolyChord running times. We recommend starting with a low number of live points and a low convergence tolerance, and build up from there towards PolyChord’s default settings (or higher, if needed).
If you would like to find the MAP (maximum-a-posteriori) or best fit (maximum of the likelihood within prior ranges, but ignoring prior density), you can swap the sampler (mcmc, polychord, etc) by minimize, as described in minimize sampler. As a shortcut, to run a minimizer process for the MAP without modifying your input file, you can simply do
cobaya-run [your_input_file_name.yaml] --minimize
Post-processing cosmological samples
Let’s suppose that we want to importance-reweight a Planck sample, in particular the one we just generated with the input above, with some late time LSS data from BAO. To do that, we add the new BAO likelihoods. We would also like to increase the theory code’s precision with some extra arguments: we will need to re-add it, and set the new precision parameter under extra_args (the old extra_args will be inherited, unless specifically redefined).
For his example let’s say we do not need to recompute the CMB likelihoods, so power spectra do not need to be recomputed, but we do want to add a new derived parameter.
Assuming we saved the sample at chains/planck, we need to define the following input file, which we can run with $ cobaya-run:
# Path the original sample
output: chains/planck
# Post-processing information
post:
suffix: BAO # the new sample will be called "chains\planck_post_des*"
# If we want to skip the first third of the chain as burn in
skip: 0.3
# Now let's add the DES likelihood,
# increase the precision (remember to repeat the extra_args)
# and add the new derived parameter
add:
likelihood:
sixdf_2011_bao:
sdss_dr7_mgs:
sdss_dr12_consensus_bao:
theory:
# Use *only* the theory corresponding to the original sample
classy:
extra_args:
# New precision parameter
# [option]: [value]
camb:
extra_args:
# New precision parameter
# [option]: [value]
params:
# h = H0/100. (nothing to add: CLASS/CAMB knows it)
h:
# A dynamic derived parameter (change omegam to Omega_m for classy)
# note that sigma8 itself is not recomputed unless we add+remove it
S8:
derived: 'lambda sigma8, omegam: sigma8*(omegam/0.3)**0.5'
latex: \sigma_8 (\Omega_\mathrm{m}/0.3)^{0.5}
Comparison with CosmoMC/GetDist conventions
In CosmoMC, uniform priors are defined with unit density, whereas in Cobaya their density is the inverse of their range, so that they integrate to 1. Because of this, the value of CosmoMC posteriors is different from Cobaya’s. In fact, CosmoMC (and GetDist) call its posterior log-likelihood, and it consists of the sum of the individual data log-likelihoods and the non-flat log-priors (which also do not necessarily have the same normalisation as in Cobaya). So the comparison of posterior values is non-trivial. But values of particular likelihoods (chi2__[likelihood_name] in Cobaya) should be almost exactly equal in Cobaya and CosmoMC at equal cosmological parameter values.
Regarding minimizer runs, Cobaya produces both a [prefix].minimum.txt file following the same conventions as the output chains, and also a legacy [prefix].minimum file (no .txt extension) similar to CosmoMC’s for GetDist compatibility, following the conventions described above.
Getting help and bibliography for a component
If you want to get the available options with their default values for a given component, use
$ cobaya-doc [component_name]
The output will be YAML-compatible by default, and Python-compatible if passed a -p / --python flag.
Call $ cobaya-doc with no arguments to get a list of all available components of all kinds.
If you would like to cite the results of a run in a paper, you would need citations for all the different parts of the process. In the example above that would be this very sampling framework, the MCMC sampler, the CAMB or CLASS cosmological code and the Planck 2018 likelihoods.
The bibtex for those citations, along with a short text snippet for each element, can be easily obtained and saved to some output_file.tex with
$ cobaya-bib [your_input_file_name.yaml] > output_file.tex
You can pass multiple input files this way, or even a (list of) component name(s).
You can also do this interactively, by passing your input info, as a python dictionary, to the function get_bib_info():
from cobaya.bib import get_bib_info
get_bib_info(info)
Note
Both defaults and bibliography are available in the GUI (menu Show defaults and bibliography for a component ...).
Bibliography for preset input files is displayed in the bibliography tab.